2022 marks 20 years since I started my MSc in computer science degree at the University of Aberystwyth. So, I thought I would take a look back at my thesis for the degree and see how far I have come since then.
The title of my thesis was "Using The Java3D API To Visualise Molecular Compounds". I used a system called Java3D to create ball and stick models of molecules. A second requirement was to allow the application to compare two molecules together.
The course I did was known as a conversion course and was designed to compress the three years of the undergraduate computer science degree into a single year, along with an introduction to Java programming bootcamp the summer before the course started. This was a year of hard work with 12 hour days and weekends of constant study. The drop out rate for the course was about 80%, with most of that being in the first few weeks.
I have been programming as a career ever since graduating, although most of my knowledge and experience has been gained outside of the degree. My degree actually focused more on desktop application development, with only a little bit about servers or websites. I covered things like data structures, sorting algorithms, database design, assembly programming, AI, networking theory and all that kind of stuff.
I certainly didn't cover PHP, JavaScript or any other web technologies during my degree (with the exception of network theory) and only picked that up afterwards.
If you don't remember 2002 (or weren't around at the time) then you need to remember that the web was very different to what it is today. Amazon mostly sold books and were just starting to sell CDs; sites YouTube and FaceBook were a couple of years away from being created and StackOverflow wasn't going to be a thing for another 6 years. As there wasn't the community of experts that there is now, and not many people were blogging highly technical subjects. Most learning was done through books or lectures. In other words, there wasn't the wealth of easily accessible knowledge that there is now.
Some Backstory
Before delving into the thesis, I think a bit of backstory is needed about how I got into the degree and how I chose this project.
I originally graduated from the University of Aberystwyth in 2000 with a degree in Microbiology, after which I got a job performing quality control on surgical implants made out of pig skin. It sounds a bit gruesome, but the implants looked like strips of chewing gum. The company doing this was essentially a startup so the processed and procedures were being built as I joined but we were soon up and running.
My skills as a microbiologist were useful as the implants needed to be handled aseptically (i.e. cleanly). Part of this work included logging the samples into a computer system and as keyboards couldn't be touched to log the samples (due to the potential of contamination) we instead logged the samples using a voice controlled system. It had a simple interface where we would speak phrases like "log pass" to record a sample of a particular size and a few other commands.
Remember that this was a good decade before voice controlled systems like Alexa where widespread, so although it was controlled via voice it was quite unreliable and often took several attempts to record things correctly. I remember that the system underwent a few designs so that you never said the same things twice during the inspection process in order to prevent false passes.
Under the hood was a MySQL server and an application written in Visual Basic 6. I know it was written in this at the start since in order to run the application we started Visual Basic 6 studio (with the raw source code on screen) and hit the "run" button. Yes, this sounds just as dodgy as it is. I did, however, get more involved with the system and ended up doing some database management and debugging tasks to aid in the development of the system.
As I got more involved with this computer system I thought I might augment things by going to night school and learning more about Visual Basic 6. So I enrolled at South Cheshire College to do some night school classes in programming around that language. I found it incredibly easy and was quickly creating applications that were above and beyond the solutions required in the assignments.
It was about this time that I decided that maybe a career change was in order and started looking at junior programming positions. I was visiting Aberystwyth a few times a year to visit friends there and when I looked into computer science degrees there I found the MSc in Computer Science course, which I applied for and quit my job in the clean room.
When I enrolled I had a 4 week intensive course on Java programming, which was hard work and often had me in the computer labs until 8pm every day. I was getting used to Visual Basic 6 and procedural programming, and Java was something completely different. I vividly remember many conversations with professors and workshop tutors trying to teach me what an "object" was. Some of the analogies where horribly tortured and just ended up confusing matters.
Going onto the course properly, I found I was getting great marks in assignments and comfortably passed all of my exams. I also met some great people on the course and got along well with all of them, which really helps the experience of the course.
After finishing my lectures I then went onto pick a thesis. I'm not sure how much things have changed in recent years, but back then we were told to pick a something from a given list of projects and supervisors. I went to a few meetings with professors who would be supervising the thesis and picked something that fitted in with my background, was interesting, and with a professor I got along with. My background in Microbiology meant that I had some background knowledge in organic chemistry, which was the main goal of the visualisation tool.
I spent the summer programming the tool and writing the 20,000 words of the thesis itself. I also had a part time job during this time so I spent pretty much most of my spare time doing all this work.
The Molecular Visualisation Tool
The program itself was written in Java 1.4 and used the Java3D package to provide the 3D component. Support for Java 1.4 ended in 2008 and Java3D hasn't been in active development for at least a decade now so the code is a little bit old.
I did spend a few hours with a Windows system trying to install older versions of Java and Java3D, but I was unable to get the system running again. I can remember when developing the program that I spent hours trying to get things installed correctly on my own machine. For some reason, Java and Java3D were installed on all the university computers, so I didn't need to worry about the requirements of the system being used to mark the project.
Thankfully I do have a few screenshots of the finished program.
This is a test molecule that I used to test the chirality of Carbon and a few other atoms consisting of Hydrogen, Oxygen, Fluorine and Lithium. The little arrow structure in the top left hand side is an axis that helped with determining the orientation of the molecule.
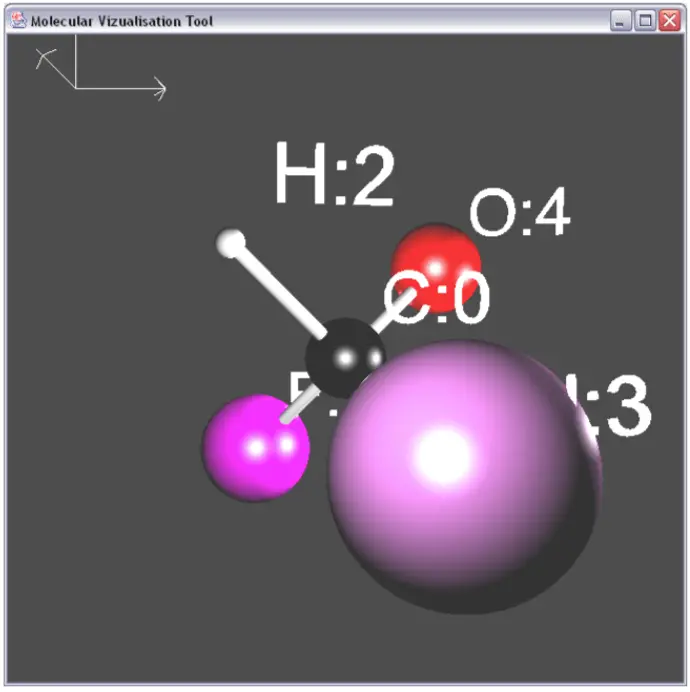
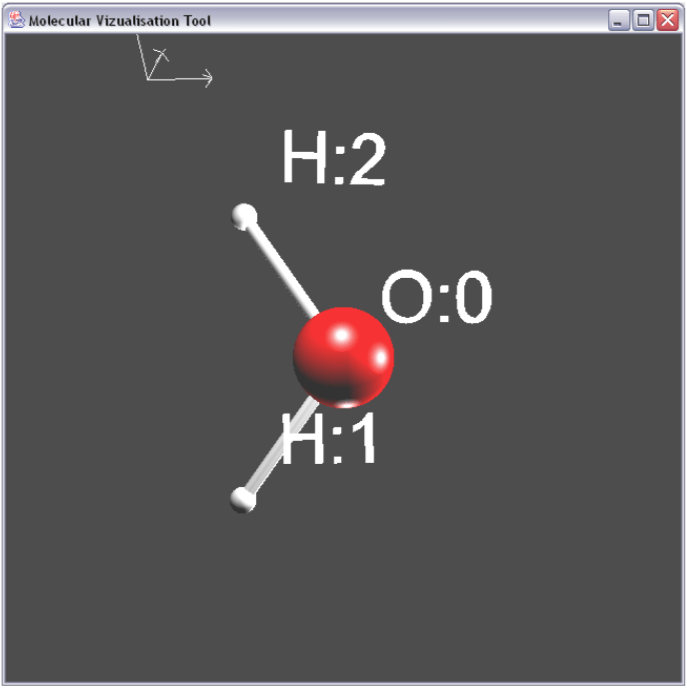
Creating a well known molecule like Water showed how well the system worked. It's easy to see the polar nature of the molecule here with one Oxygen and two Hydrogen molecules.
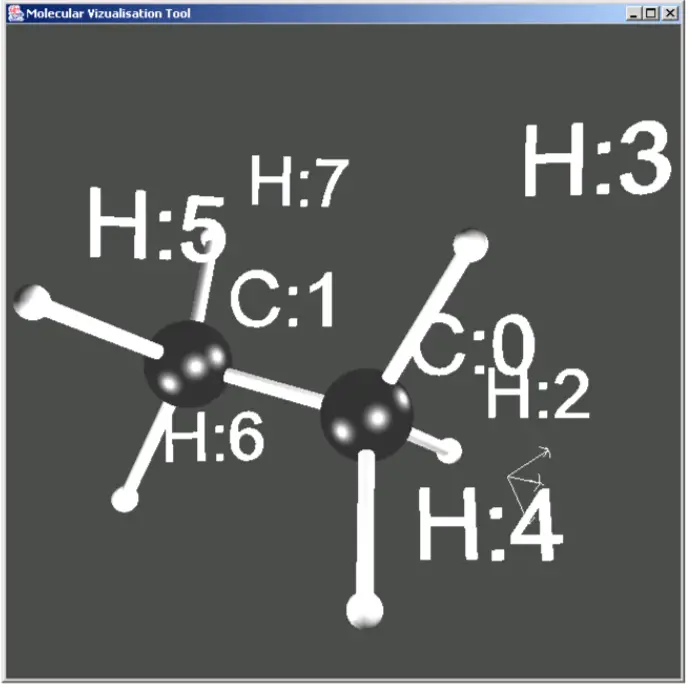
More complex molecules like ethene were also possible. Ethene consists of two Carbon atoms and 6 Hydrogen atoms.
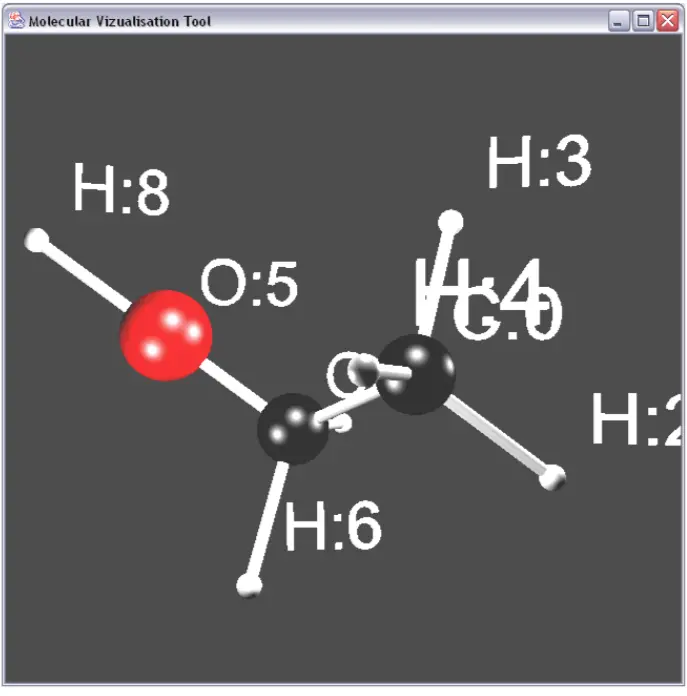
Taking that a step further, here is a molecule of ethanol with the chemical structure of C2H6O.
The above scenes were not static. I also added mouse navigation that allows a user to rotate the molecule and zoom in and our of the created 3D image.
This was phase one of the program. Phase two consisted of three different windows that represented two molecules and a window for animating the difference between the two molecules. I don't have any screenshots of this in action unfortunately.
The program itself was pretty simple with little or no interface. In fact, changing molecules consisted of copying and pasting a bunch of atom generation code into a section of the program.
Before delving into the code, let's take a quick look at the structure of the program.
Structure
The structure of the application was created in (roughly) two parts with a data structure and a graphical interface.
The data structure of the molecules was contained in a graph, which was created using an array of linked lists. This allowed arbitrary numbers of elements to be created in order to store Atom objects, which was all managed by a class called Molecule. The Atom objects were the cornerstone of the data structure as they stored data about the element, including how many bonds could be created.
The graphics of the program was setup using Java's Swing package, which was commonly used for user interfaces at the time (and might still be, I haven't looked!). A simple GUI interface was created that then had the Java3D scene injected into it. A class called MoleculeTo3d was used to translate the Molecule class into the ball and stick models using standard Java3D Sphere and Cylinder objects. As you can see from the screenshots I also added in some text to show the element name and the ID of the Atom around each Sphere object in the scene.
In order to present a decent amount of light in the Java3D scene I added both ambient light as well as three spotlights to give some directional light to the objects. You can see these direction lights in the screenshots above as each atom sphere had three white dots on it.
There were a lot of smaller components involve in the structure of the application. I found the following diagram showing this structure.
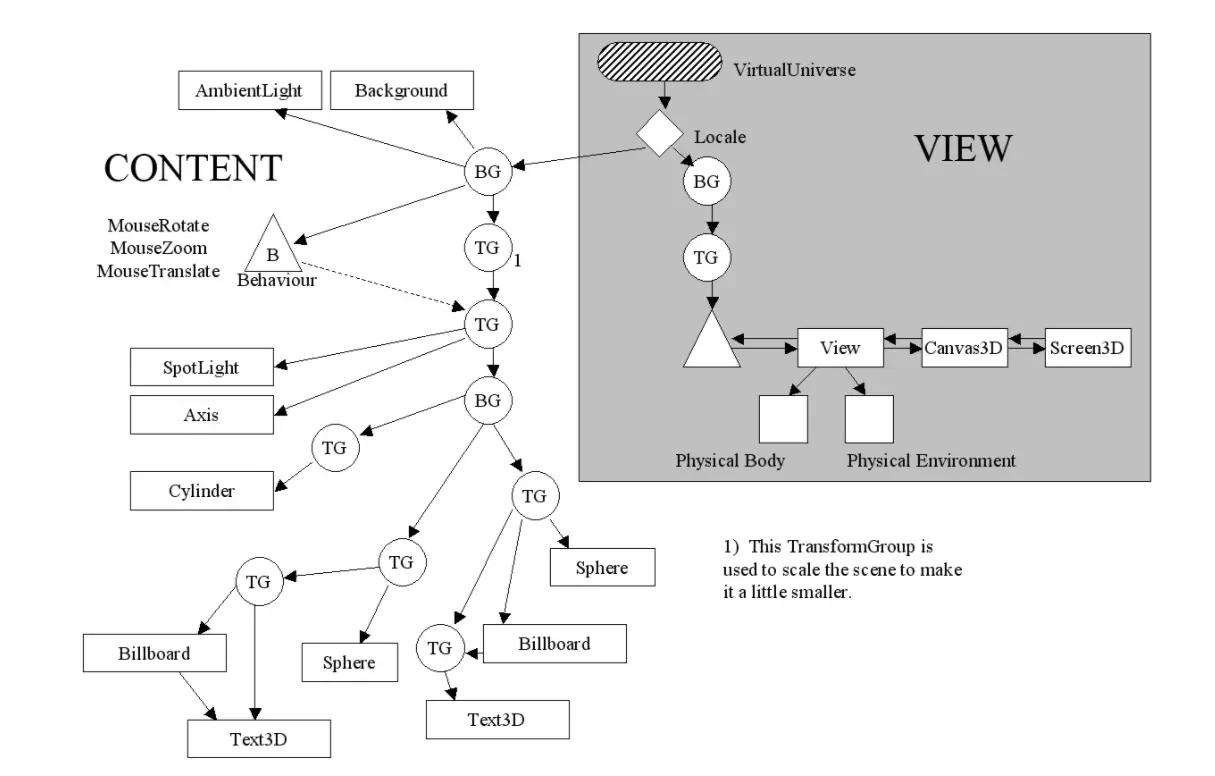
In the diagram TG stands for TransformGroup and BG stands for BranchGroup. A TransformGroup is a way of moving a collection of objects either by rotating them around a point or by moving them in space. A BranchGroup is just a way of grouping a bunch of objects together so that they can be treated as one "thing". In the diagram above you can see that I add the Atom shapes under a BranchGroup and use TransformGroups to move the cylinders and spheres around the scene.
The separation of Content and View is essentially my way of showing the difference between my ball and stick models and the camera used to view the scene.
This shows that I clearly understood things about the way in which a 3D scene was generated.
Code
The code of the project consists of 16 files and around 3,500 lines of code.
The code I wrote for the graph data structure is actually ok. It's well written and contains a number of small methods that control different aspects of the linked lists. Things get a bit messy in the Molecule class since that class is acting as the graph management class and also includes the different molecule management features. I think a better approach would have been to abstract the graph entirely away from the molecule so that the complexity is hidden from the Molecule class.
Where things get a really messy is in the translation of the Molecule structure to the 3D structure. The MoleculeTo3d class does some of the work in terms of creating groups of spheres and cylinders, but the SwingSceneContent class (used to generate the interface) also has a lot of logic regarding this process. The molecule is set up in the SwingSceneContent class and partially translated in that class and the MoleculeTo3d class.
There isn't really a user interface outside of the Java3D frame that the SwingSceneContent class creates, so the molecule is built up using the swing interface class. This means that all of the creation of atoms and bonds is baked right into the scene creation code.
As an example, here is the code involved in setting up a molecule of ethanol with the different Atom objects being created and connected together to form the structure of the molecule.
public Molecule setUpMolecule(){
////////////////////////////
// start of atom creation //
////////////////////////////
// create atom objects
Atom atom0 = new Atom("C");
Atom atom1 = new Atom("C");
Atom atom2 = new Atom("H");
Atom atom3 = new Atom("H");
Atom atom4 = new Atom("H");
Atom atom5 = new Atom("O");
Atom atom6 = new Atom("H");
Atom atom7 = new Atom("H");
Atom atom8 = new Atom("H");
// create Molecule object
Molecule mol = new Molecule(count);
// add Atom objects to Molecule object
mol.addAtom(atom0);
mol.addAtom(atom1);
mol.addAtom(atom2);
mol.addAtom(atom3);
mol.addAtom(atom4);
mol.addAtom(atom5);
mol.addAtom(atom6);
mol.addAtom(atom7);
mol.addAtom(atom8);
// add Bond objects to Atom objects in the Molecule object
mol.addBond(atom0,atom1,1);
mol.addBond(atom0,atom2,1);
mol.addBond(atom0,atom3,1);
mol.addBond(atom0,atom4,1);
mol.addBond(atom1,atom5,1);
mol.addBond(atom1,atom6,1);
mol.addBond(atom1,atom7,1);
mol.addBond(atom5,atom8,1);
//////////////////////////
// end of atom creation //
//////////////////////////
mol.setCurrentAtom(mol.atoms[0].getFirst());
mol.setCurrentBond(mol.bonds.getFirst());
return mol;
}
I had also provided a few example molecules that could be copied and pasted into this section to generate different molecules. I don't know if the person marking the project bothered to do this. I wouldn't have bothered if I saw this handed to me!
Whilst I have tried to separate concerns in the system I've ultimately not abstracted things enough and there is significant overlap between classes. There's no clear line where you can say "the 3D scene starts here".
Perhaps most confusingly, the Atom class is never extended to create different types of element. The class itself translates the element into attributes for other classes to work on. In the Atom class there is a method called elementAttributes() that is called from the constructor that demonstrates this.
private void elementAttributes(){
if(getElementSymbol() == "C"){
setElementName("Carbon");
setAtomicNumber(6);
setAtomicRadius(0.6f);
setValency(4);
setHydrogenCount(4);
}else if(getElementSymbol() == "N"){
setElementName("Nitrogen");
setAtomicNumber(7);
setAtomicRadius(0.7f);
setValency(3);
setHydrogenCount(3);
}else if(getElementSymbol() == "O"){
This if statement goes on for over 60 lines of code! Oh dear...
When translating this into sphere objects this resulted in a similarly lengthy if statement.
public BranchGroup createAtom(Atom theAtom){
BranchGroup atomGroup = new BranchGroup();
theAtom.setAtomDisplayed(true);
if(theAtom.getElementSymbol() == "C"){
////////////
// CARBON //
////////////
Sphere carbon = new Sphere((float)theAtom.getAtomicRadius(),Sphere.GENERATE_NORMALS,200,setTheAppearance(0.0f,0.0f,0.0f));
atomGroup.addChild(carbon);
}else if(theAtom.getElementSymbol() == "O"){
////////////
// OXYGEN //
////////////
Sphere oxygen = new Sphere(theAtom.getAtomicRadius(),Sphere.GENERATE_NORMALS,200,setTheAppearance(1.0f,0.0f,0.0f));
atomGroup.addChild(oxygen);
}
This continues for another 60 lines as well.
The bond type in the Bond class does a similar thing. I should note that I never actually coded in the 3D shape for aromatic or borane bonds, but I still added the functionality.
public void setBondType(int theBondType){
if(theBondType == SINGLE){
bondType = SINGLE;
}else if(theBondType == DOUBLE){
bondType = DOUBLE;
}else if(theBondType == TRIPLE){
bondType = TRIPLE;
}else if(theBondType == AROMATIC){
bondType = AROMATIC;
}else if(theBondType == BORANE){
bondType = BORANE;
}else if(theBondType == DEFAULT){
bondType = SINGLE;
}else if(bondType == BREAK){
bondType = BREAK;
}else if(bondType == ERROR){
bondType = SINGLE;
}
}
I'm sure I knew something about inheritance at this point as I have done it at least once in the project. For some reason I decided not to use it when creating things that obviously called for it. Doing things without inheritance meant writing lots and lots of code to build up the picture of each type of atom.
If you're wondering why the formatting looks a little strange in the above examples then it's because I had used both tabs and spaces in the project and they don't translate correctly into the code viewer. In fact, the entire project uses tabs and spaces randomly and interchangeably, even on the same line. Far from settling that debate I have just ignored it and coded randomly.
One good thing is that each class has a number of simple checks in the "public static void main()" functions that I added in an effort to ensure that the code ran correctly. This means that if I ran the compiled class on its own it would print out some test results. This is an attempt at creating unit tests without ever knowing what unit tests were. These functions perform things like adding items to a list and ensuring that the length of the list is the same as the number of items added.
What surprised me was the amount of commented out code there was. There are the comments that show what is going on in the code, but there were also signposts around the code that just showed what type of thing was being created at the time.
There are also about 40 instances of the following snippet of code with the System.out.println() statement randomly being enabled.
//DEBUG
//System.out.println("SwingSceneContent:createSceneGraph");
I was clearly using print statements to debug and add logging without having an interactive debugger.
Conclusion
This code clearly shows that although I knew what an object is I really didn't understand much about inheritance or abstraction using objects. I could have saved myself a lot of code by abstracting things into classes of certain types. Quite a few functions run on for hundreds of lines of code (with lots and lots of nested if statements) and I think that's partially due to this lack of abstraction.
What is clear is that business logic is embedded in code that doesn't need it. I found lots of boilerplate code that sets things up mixed in with logic that translates molecules into a collection of spheres and cylinders. For example, the code that adds sphere objects to the scene is in the same block of code as adding lights sources.
I seem to remember spending hours and hours creating little test programs that showed things like rotation or how to create a sphere. This was done to learn how to use Java3D and about 3D mathematics, but I still find myself doing this sort of prototyping to learn things about new systems or languages. Doing so is a good way of getting familiar with the key concepts and then having a collection of examples to look back at.
Ultimately, the program worked quite well to produce simple molecules, but failed when presented with more complicated structures. I remember not being able to solve an issue that caused long chain Carbon molecules to fold on themselves and create a mess. Aromatic rings (a staple of organic chemistry) were also not possible as I hadn't solved that initial problem and couldn't get the rings to create correctly.
The support I received from my professor during this period was almost non existent. We had weekly meetings where we went over progress, and although I asked for help I never received any more than abstract guidance. I remember getting frustrated at the folding problem I mentioned before, and sending my project over asking for some help. My professor admitted not to even have looked at the code, and I left having to figure out things for myself.
I remember other people on my course getting very frustrated by their projects as they spent weeks writing pages and pages of plans but never writing any code. My professors guidance was to "get stuck in" and gave me the idea of the graph data structure which I immediately started creating. In retrospect I should have spent more time planning out how things would work as that would have prevented me from coding myself into a broken corner.
Originally, I was also meant to have an interface in the program that allowed users to enter Simplified Molecular-Input Line-Entry System (SMILIES) text so that the user didn't have to do any copy and pasting to change molecules. For example, the user would enter CC for ethene (the hydrogen atoms are assumed to be there).
I added a text box to accept the information for SMILIES format strings, but couldn't figure out how to translate the text into molecules and so removed it. This is the reason why the interface is so lacking and I talked about doing this as a future improvement in the thesis document.
All of the code was written using a code editor called GEdit, which only did basic code syntax highlighting and find/replace functions. I didn't have modern tools like inline debugging, auto-formatting, code completion, static analysis or any of the other things that I rely on to code on a day to day basis. This is essentially a step above notepad in terms of an interface to write code and meant many hours more of adding debug statements or trying to apply abstract API documentation into concrete code. I think that added to the amount of time spent just fiddling with things. I think this is probably the reason for tabs and spaces being used randomly.
This was also a few years before I found source control, and I think that shows in the project a little. I would get something working and then not want to break it in case I couldn't get back to the original state. Having the safety net of source control is useful in my current career as I can experiment with things.
I think, however, that I managed to create quite a lot in just a few of months of work. I essentially learnt a new API in Java3D along with a lot of complex 3D maths, created a complex data structure, coded a graphics engine and wrote the 20,000 words of the thesis. I was quite proud of what I had created and it got me quite a good mark. I haven't touched Java since leaving university, but I don't really miss it.
If I were to sit down and create the same program again I would change quite a few things around, but probably keep some of the key components in place. Creating more abstraction of things would be my first step so that it would be easier to see how things fitted together, rather than have colossal "if" statements. Massive control structures like this are difficult to debug and maintain (trust me!). Passing objects as parameters would be much more common as well since lots of things are baked into the code in this project.
My top tip if you are getting into object oriented programming would be to look at how objects fit together to solve problems. Get beyond concepts like factories and interfaces and look at how inheritance can be used to prevent code reuse. Having that knowledge helps you plan your code out before you write it.
Also, if you are feeling like a career change then I say go for it! In reality I started my path to computer science a little later than some people, but I don't think that matters. Doing another degree probably necessary these days, but my first degree taught me how to study and how to write for coursework, which were essential skills for doing an MSc.






Add new comment